Genome size is the most fundamental property of a genome and varies widely across the tree of life in a vast spectrum spanning several orders of magnitude. Despite great efforts, the mechanism of genome size variation remains an aspect of concerns in population genetics and comparative genomics. To date, several hypotheses have been raised but invoked extensive controversy. In 2003, Lynch and Conery proposed the famous hypothesis that evolution of genome complexity was driven mainly by non-adaptive stochastic forces, which is strongly supported by the observation that genome size is negatively correlated with effective population size (Ne).
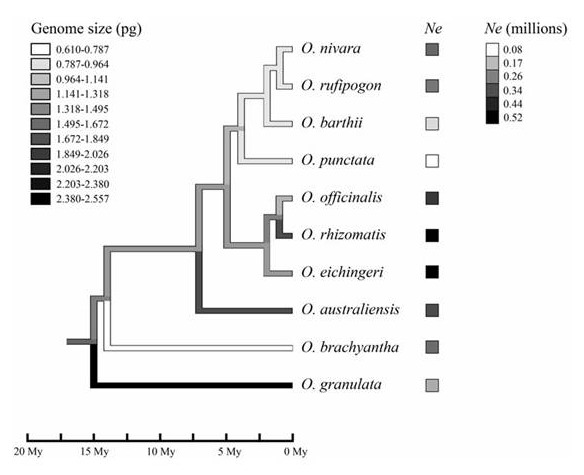
The genusOryza consists of two cultivated and approximately 22 wild species distributed across the world. With the fully resolved phylogeny and fourfold genome size variation in diploid species, the Oryza genus provides an ideal system to study the mechanism of genome size variation. Dr. GE Song’s group from State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, CAS, used sequences of 26 nuclear gene fragments for 10 species representing all diploid genome types in Oryza to investigate the potential mechanism of genome size variation in this genus. By pairwise correlation analyses, they did not detect a negative relationship between Ne and genome size despite about 6.5-fold interspecies Ne variation. After applying phylogenetically independent contrasts for Ne, they repeated correlation analysis and found no correlation between Ne and genome size either. These observations suggest that the genome size variationin the Oryza species cannot be explained simply by the effect of effective population size. Either, mating system and other hypotheses including adaptive and neutral theories cannot account for the variation.
This study is the first to investigate the mechanism of genome size variation based on a fine-scale comparison among closely related species, and provides a well-studied case for the future research. The above results have been published in Evolution (2012, 66:3302-3310). This work was financially supported by the National Basic Research Program of China and the National Natural Science Foundation of China.
To see the paper, please click: http://onlinelibrary.wiley.com/doi/10.1111/j.1558-5646.2012.01674.x/abstract
For more information, please contact Professor GE Song at: gesong@ibcas.ac.cn
Genome size is the most fundamental property of a genome and varies widely across the tree of life in a vast spectrum spanning several orders of magnitude. Despite great efforts, the mechanism of genome size variation remains an aspect of concerns in population genetics and comparative genomics. To date, several hypotheses have been raised but invoked extensive controversy. In 2003, Lynch and Conery proposed the famous hypothesis that evolution of genome complexity was driven mainly by non-adaptive stochastic forces, which is strongly supported by the observation that genome size is negatively correlated with effective population size (Ne).
The genusOryza consists of two cultivated and approximately 22 wild species distributed across the world. With the fully resolved phylogeny and fourfold genome size variation in diploid species, the Oryza genus provides an ideal system to study the mechanism of genome size variation. Dr. GE Song’s group from State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, CAS, used sequences of 26 nuclear gene fragments for 10 species representing all diploid genome types in Oryza to investigate the potential mechanism of genome size variation in this genus. By pairwise correlation analyses, they did not detect a negative relationship between Ne and genome size despite about 6.5-fold interspecies Ne variation. After applying phylogenetically independent contrasts for Ne, they repeated correlation analysis and found no correlation between Ne and genome size either. These observations suggest that the genome size variationin the Oryza species cannot be explained simply by the effect of effective population size. Either, mating system and other hypotheses including adaptive and neutral theories cannot account for the variation.
This study is the first to investigate the mechanism of genome size variation based on a fine-scale comparison among closely related species, and provides a well-studied case for the future research. The above results have been published in Evolution (2012, 66:3302-3310). This work was financially supported by the National Basic Research Program of China and the National Natural Science Foundation of China.
To see the paper, please click: http://onlinelibrary.wiley.com/doi/10.1111/j.1558-5646.2012.01674.x/abstract
For more information, please contact Professor GE Song at: gesong@ibcas.ac.cn  |
Phylogeny for 10 Oryza species with a reconstruction of ancestral genome sizes. Shades of the
branches on the tree represent the genome sizes, and shades of squares after the species
names indicate the mean Ne estimates for the species. Time scale is provided below the tree.
