Gene duplications are arguably the driving force of organismal evolution. Duplicate genes, if survived, tend to diverge in regulatory and coding regions. Coding divergences, which are usually prerequisite of functional differentiation, can be caused by nucleotide substitutions or exon-intron structural changes. Scientists have had limited knowledge in the latter case until recently, when Prof. KONG Hongzhi and his research group at the State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, the Chinese Academy of Sciences, investigated the prevalence, importance and underlying mechanisms of structural divergences during the evolution of duplicate and non-duplicate genes.
The group led by Dr. Kong analyzed 612 pairs of paralogous genes and 300 pairs of orthologous genes in model plants and found that changes in exon-intron structure have been widespread in gene evolution. They also found that divergences in exon-intron structure can be accomplished through three main types of mutations (i.e. intra-exonic insertion/deletion, exonization/pseudoexonization, and exon/intron gain/loss), each of which contributed differently to structural divergence. Notably, however, compared with paralogs with similar evolutionary times, orthologs have accumulated significantly fewer structural changes, whereas the amounts of amino acid replacements remained largely unchanged. This suggests that structural divergences have played a more important role during the evolution of duplicate rather than non-duplicate genes and have led to the generation of genes with novel functions in short times. These findings help better understand the general patterns of gene evolution and highlighted the importance of structural divergence in gene and organismal evolution. (PNAS, 2012, doi:10.1073/pnas.1109047109)
The research work was recently published online on the PNAS (http://www.pnas.org/content/109/4/1187.full.pdf+html ).
Contacts: Dr. KONG Hongzhi, hzkong@ibcas.ac.cn
The research work was recently published online on the PNAS (http://www.pnas.org/content/109/4/1187.full.pdf+html ).
Contacts: Dr. KONG Hongzhi, hzkong@ibcas.ac.cn
 |
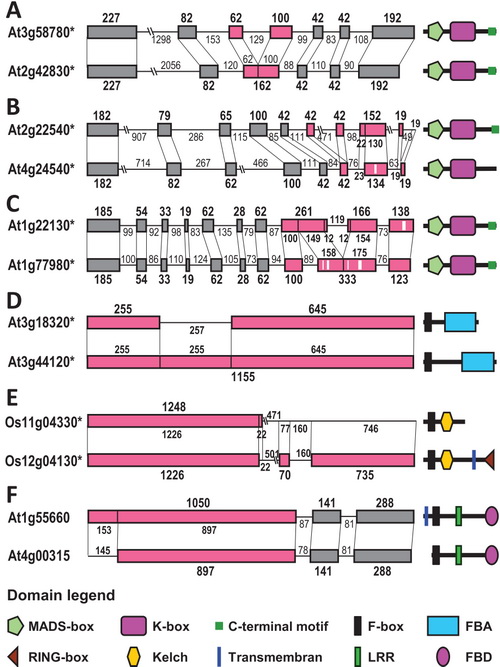
The exon–intron structures of six pairs of representative sibling
paralogs and the domain organization of their proteins, showing the three
types of underlying mechanisms for structural divergences.
