Why sweet sorghum tastes sweeter than grain sorghum? Researchers from the Institute of Botany, Chinese Academy of Sciences (IOB) compared the genome-wide variation between sweet and grain sorghum using genome sequences, and found they are differ in nearly 1,500 genes.
Sorghum (Sorghum bicolor) is the number 5 staple crop and is globally produced as a source of food, feed, fibre and fuel. Sweet sorghum is a natural variant of the grain sorghum and has several distinct features including higher stem sugar and juice accumulation, taller plants and bigger biomass production, all of ‘sweetness’ traits make sweet sorghum the crop of choice for the first and second biofuel production. Genome-wide information on the genetic variation between sweet and grain sorghum is required for dissecting the basis of these important traits and for tailor-designed breeding of this important C4 crop.
Researchers applied the next-generation sequencing technologies and resequenced two sweet and one grain sorghum inbred lines and identified a set of nearly 1,500 genes differentiating sweet and grain sorghum. These genes fall into 10 major metabolic pathways involved in sugar and starch metabolisms, lignin and coumarin biosynthesis, nucleic acid metabolism, stress responses and DNA damage repair. The effort has also uncovered 1,057,018 SNPs, 99,948 indels of 1-10bp in length and 16,487 presence/absence variations as well as 17,111 CNVs. This work is jointly finished by IOB and research teams from Temasek, Life Sciences Laboratory at Singapore and Beijing Genomics Institute at ShenZhen.
This is a first report of the identification of genome-wide patterns of genetic variationin sorghum. The leading scientist, Dr. Hai-Chun Jing, indicates that although rudimentary, the genomic resources acquired through this research including the high-density SNP and indel markers, will be a valuable for future gene - phenotype studies and the molecular breeding of this important crop and related species.
The results are reported in the paper published online on November 21, 2011 in Genome Biology (doi:10.1186/gb-2011-12-11-r114). The work is supported by a grant of the Knowledge Innovation Programme of the Chinese Academy of Sciences and the Temasek Life Sciences Laboratory, Singapore.
Click to see the full text of the article: http://genomebiology.com/content/pdf/gb-2011-12-11-r114.pdf
For more information or to set up interviews, please contact: Dr. Haichun Jing; Email: hcjing@ibcas.ac.cn ; Tel: +86- 10- 62836576
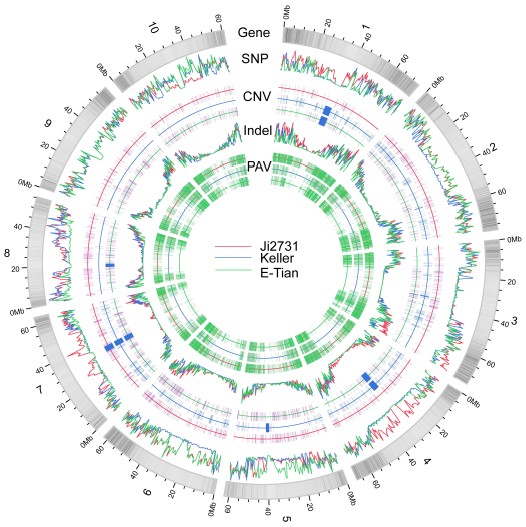
A figure deciphering the landscape of genetic variation identified in the sorghum genomes.
Why sweet sorghum tastes sweeter than grain sorghum? Researchers from the Institute of Botany, Chinese Academy of Sciences (IOB) compared the genome-wide variation between sweet and grain sorghum using genome sequences, and found they are differ in nearly 1,500 genes.
Sorghum (Sorghum bicolor) is the number 5 staple crop and is globally produced as a source of food, feed, fibre and fuel. Sweet sorghum is a natural variant of the grain sorghum and has several distinct features including higher stem sugar and juice accumulation, taller plants and bigger biomass production, all of ‘sweetness’ traits make sweet sorghum the crop of choice for the first and second biofuel production. Genome-wide information on the genetic variation between sweet and grain sorghum is required for dissecting the basis of these important traits and for tailor-designed breeding of this important C4 crop.
Researchers applied the next-generation sequencing technologies and resequenced two sweet and one grain sorghum inbred lines and identified a set of nearly 1,500 genes differentiating sweet and grain sorghum. These genes fall into 10 major metabolic pathways involved in sugar and starch metabolisms, lignin and coumarin biosynthesis, nucleic acid metabolism, stress responses and DNA damage repair. The effort has also uncovered 1,057,018 SNPs, 99,948 indels of 1-10bp in length and 16,487 presence/absence variations as well as 17,111 CNVs. This work is jointly finished by IOB and research teams from Temasek, Life Sciences Laboratory at Singapore and Beijing Genomics Institute at ShenZhen.
This is a first report of the identification of genome-wide patterns of genetic variationin sorghum. The leading scientist, Dr. Hai-Chun Jing, indicates that although rudimentary, the genomic resources acquired through this research including the high-density SNP and indel markers, will be a valuable for future gene - phenotype studies and the molecular breeding of this important crop and related species.
The results are reported in the paper published online on November 21, 2011 in Genome Biology (doi:10.1186/gb-2011-12-11-r114). The work is supported by a grant of the Knowledge Innovation Programme of the Chinese Academy of Sciences and the Temasek Life Sciences Laboratory, Singapore.
Click to see the full text of the article: http://genomebiology.com/content/pdf/gb-2011-12-11-r114.pdf
For more information or to set up interviews, please contact: Dr. Haichun Jing; Email: hcjing@ibcas.ac.cn ; Tel: +86- 10- 62836576
 |
| A figure deciphering the landscape of genetic variation identified in the sorghum genomes. |
(image by IOB)
