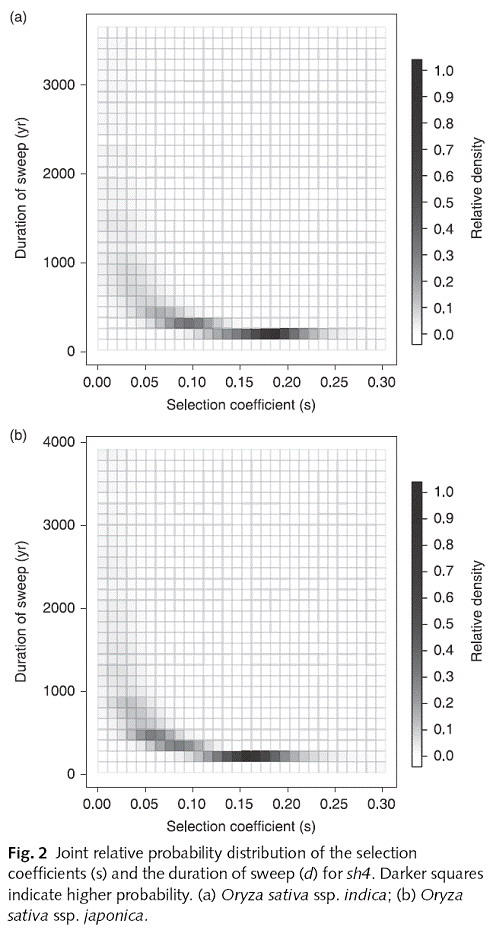
Molecular cloning of major quantitative trait loci (QTLs) responsible for the reduction of rice grain shattering, a hallmark of cereal domestication, provided opportunities for in-depth investigation of domestication processes. Here, we studied nucleotide variation at the shattering loci, sh4 and qSH1, for cultivated rice, Oryza sativa ssp. indica and Oryza sativa ssp. japonica, and the wild progenitors, Oryza nivara andOryza rufipogon. The nonshattering sh4 allele was fixed in all rice cultivars, with levels of sequence polymorphism significantly reduced in both indica and japonica cultivars relative to the wild progenitors. The sh4 phylogeny together with the neutrality tests and coalescent simulations suggested that sh4 had a single origin and was fixed by artificial selection during the domestication of rice. Selection on qSH1 was not detected in indica and remained unclear in japonica. Selection on sh4 could be strong enough to have driven its fixation in a population of cultivated rice within a period of c. 100 yr. The slow fixation of the nonshattering phenotype observed at the archeological sites might be a result of relatively weak selection on mutations other than sh4 in early rice cultivation. The fixation of sh4 could have been achieved later through strong selection for the optimal phenotype.
New Phytol. 2009 Nov;184(3):708-20. Epub 2009 Aug 5.
Zhang LB, Zhu Q, Wu ZQ, Ross-Ibarra J, Gaut BS, Ge S, Sang T.
State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China.