After having reported heterogeneity in the evolutionary rates of SKP1 -like genes in 2004, Prof. Hongzhi Kong and his colleagues further investigated the gene structure, chromosomal location and phylogenetic relationships of plant SKP1 genes. They found that the evolution of SKP1 genes in angiosperms was a rapid "birth-and-death" process, with the birth rate more than 10 times the average rate of gene duplication and even higher than that of other rapidly duplicating plant genes, such as type I MADS box genes, R genes, and genes encoding receptor-like kinases. Further analyses suggested that a relatively large proportion of the duplication events may be explained by tandem duplication and retroposition, but few, if any, were likely to be due to segmental duplication. This study not only clarified the evolutionary history of the SKP1 gene family, but also represents a thorough investigation showing that retroposition can play an important role in the evolution of a gene family whose members do not encode mobile elements ( Plant J , 2007 ).
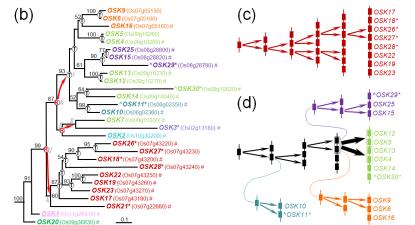
Figure. Evolution of the Arabidopsis SKP1 -like genes ( ASKs ). (a) Chromosomal locations and (b) phylogenetic relationships. The orientations of each gene are shown by arrows. Locus names are shown in parentheses. Genes with the specific intron are underlined, while those with different, late-gained introns are marked with a ‘^'. Expressed genes are labeled with a ‘#' . Real pseudogenes (i.e. genes with partial sequences) are highlighted with a ‘w', and putative pseudogenes are labeled with an asterisk (*). The letters T, S and R on the nodes of the phylogenetic tree indicate the positions where tandem duplication, segmental duplication and retroposition have occurred, respectively. Hypothesized donor genes and retrogenes during retroposition are connected by curved arrows.
