The Hippo signaling pathway plays important roles in organ size control and tumorigenesis in animals. Mutations in core components of the pathway often cause over-growth of organs or tumor. The pathway is highly conserved in animals, but it is not clear whether it also exists and functions in plants. Prof. CHENG Youfa’s team at the Institute of Botany, Chinese Academy of Sciences found that a key component in the Hippo pathway, NCP1/AtMOB1A, plays critical roles in auxin-mediated plant development. By using genetics and biochemistry approaches, they demonstrated that NCP1/AtMOB1A synergistically interacts with key genes in auxin biosynthesis, polar transport and signal transduction to regulate organogenesis and embryogenesis in Arabidopsis. AtMOB1A is highly homologous to animal MOB1 proteins. Interestingly, expression of the Drosophila MOB1 gene Mats, driven by the NCP1/AtMOB1A promoter, in Arabidopsis atmob1a mutant was able to rescue its developmental defects. These findings indicate that MOB1 proteins are functionally conserved in animals and plants. This work identified a new player in auxin-mediated plant development, and opens up novel perspectives about the mechanisms by which auxin regulates organogenesis. This work entitled “NCP1/AtMOB1A plays key roles in auxin-mediated Arabidopsis development” is published online in PLOS Genetics (DOI: 10.1371/journal.pgen.1005923) on March 4th, 2016. CUI Xiaona and GUO Zhiai are the co-first authors. SONG lizhen and WANG Yanli also contributed to the study. This study was supported by the grants from the National Basic Research Program of China, and the National Natural Science Foundation of China.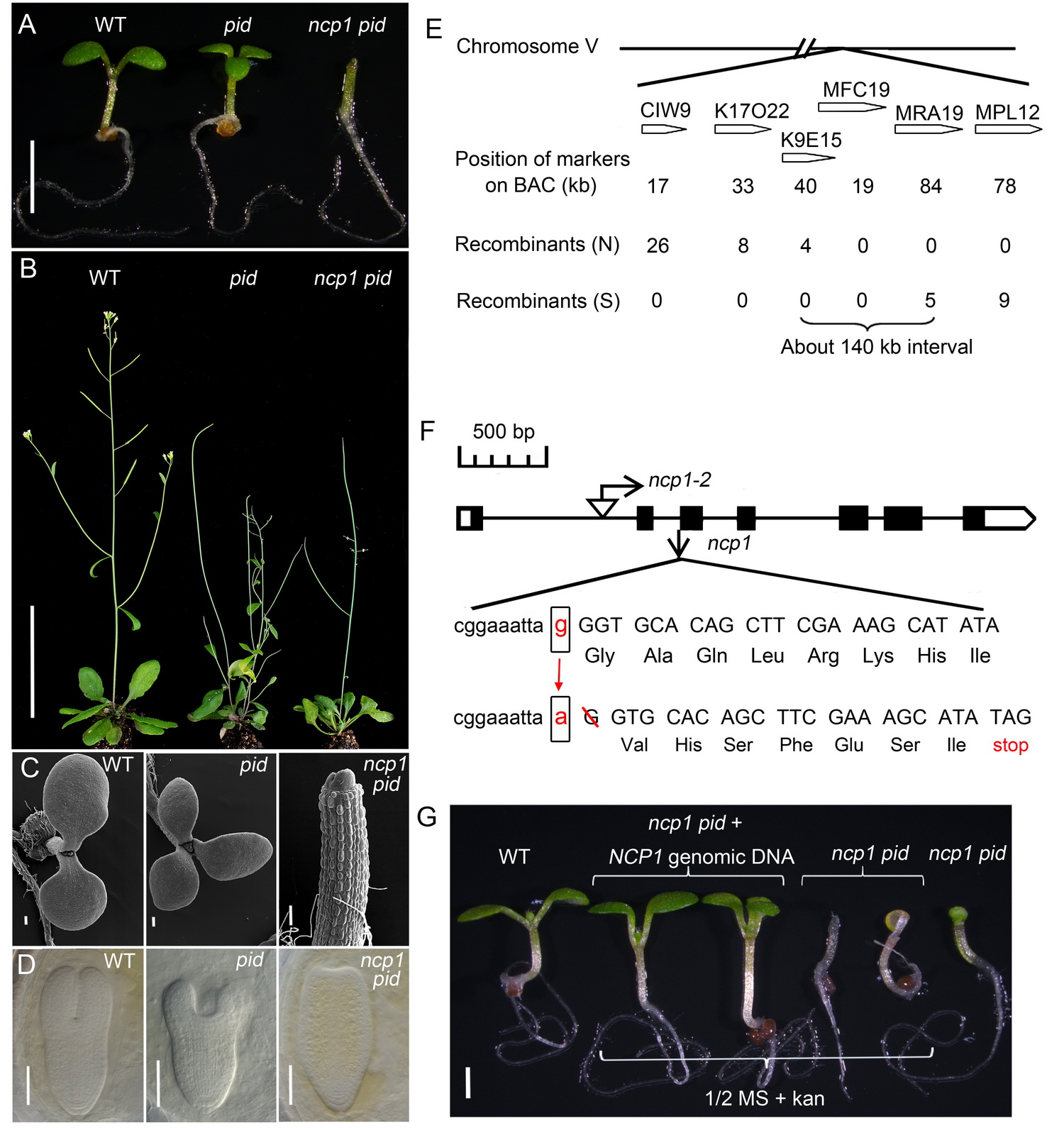
Fig 1. Identification and molecular cloning of ncp1. A-D, ncp1 enhanced pid developmental defects. The double mutants displayed no-cotyledon phenotype during embryogenesis. E-G, Map-based cloning of NCP1 and complementation.

Fig 2. The Drosophila MOB1 gene Mats can functionally substitute for NCP1. A-B, Expression of Mats in ncp1 driven by NCP1 promoter fully rescued developmental defects of ncp1. C-D, Rescue of silique length and ovule number.
Link:
http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005923
Contact information:
Prof. CHENG Youfa
Key Laboratory of Plant Molecular Physiology
Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China
e-mail: yfcheng@ibcas.ac.cn
http://www.klpmp.net/en/groups_display.asp?ktid=52&ktclass=2

Fig 1. Identification and molecular cloning of ncp1. A-D, ncp1 enhanced pid developmental defects. The double mutants displayed no-cotyledon phenotype during embryogenesis. E-G, Map-based cloning of NCP1 and complementation.

Fig 2. The Drosophila MOB1 gene Mats can functionally substitute for NCP1. A-B, Expression of Mats in ncp1 driven by NCP1 promoter fully rescued developmental defects of ncp1. C-D, Rescue of silique length and ovule number.
Link:
http://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1005923
Contact information:
Prof. CHENG Youfa
Key Laboratory of Plant Molecular Physiology
Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China
e-mail: yfcheng@ibcas.ac.cn
http://www.klpmp.net/en/groups_display.asp?ktid=52&ktclass=2
