Gene duplication is the primary source of new genes and novel functions. A particular challenge is to understand the molecular mechanisms responsible for the initial retention and subsequent divergence of newly created duplicate genes. Recently, Dr. Qing-Yin Zeng and his collaborators from the Institute of Botany, Chinese Academy of Sciences, found that subcellular relocalization and positive selection play key roles in the retention of duplicate genes. The class III peroxidases (PRXs) are plant-specific enzymes that play important roles in cell wall metabolism and in response to biotic and abiotic stresses. Dr. Qing-Yin Zeng’s group reconstructed the evolutionary history of PRX genes from the Populus trichocarpa genome. They found that two large tandem-arrayed clusters of PRXs evolved from an ancestral cell wall type PRX to vacuole type, followed by tandem duplications and subsequent functional specification. Substitution models identified seven positively selected sites in the vacuole PRXs. These positively selected sites showed significant effects on the biochemical functions of the enzymes.
This work has been published in The Plant Cell (Ren et al., 2014, Plant Cell, 26: 2404-2419), and highlighted by Plant Cell “In Brief” article (Jennifer Mach, 2014, When to hold them: Retention of duplicate genes in poplar, Plant Cell, 26: 2283). Also this work has been recommended by FACULTY of 1000 (http://f1000.com/prime/718453323)
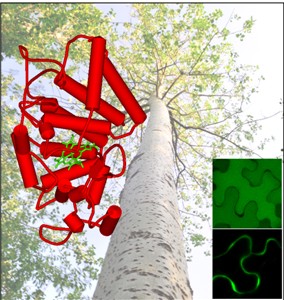
Figure. The structure and subcellular localizations of Populus PRX proteins
Gene duplication is the primary source of new genes and novel functions. A particular challenge is to understand the molecular mechanisms responsible for the initial retention and subsequent divergence of newly created duplicate genes. Recently, Dr. Qing-Yin Zeng and his collaborators from the Institute of Botany, Chinese Academy of Sciences, found that subcellular relocalization and positive selection play key roles in the retention of duplicate genes. The class III peroxidases (PRXs) are plant-specific enzymes that play important roles in cell wall metabolism and in response to biotic and abiotic stresses. Dr. Qing-Yin Zeng’s group reconstructed the evolutionary history of PRX genes from the Populus trichocarpa genome. They found that two large tandem-arrayed clusters of PRXs evolved from an ancestral cell wall type PRX to vacuole type, followed by tandem duplications and subsequent functional specification. Substitution models identified seven positively selected sites in the vacuole PRXs. These positively selected sites showed significant effects on the biochemical functions of the enzymes.
This work has been published in The Plant Cell (Ren et al., 2014, Plant Cell, 26: 2404-2419), and highlighted by Plant Cell “In Brief” article (Jennifer Mach, 2014, When to hold them: Retention of duplicate genes in poplar, Plant Cell, 26: 2283). Also this work has been recommended by FACULTY of 1000 (http://f1000.com/prime/718453323)

Figure. The structure and subcellular localizations of Populus PRX proteins
